Traditionally, culture-based techniques have predominated when testing for bacteria. If an organism can grow in vitro, then it is viable and may constitute a threat to health or to the integrity of a product. Yet not all viable bacteria are currently capable of being cultured, and even if they cannot replicate on artificial media, they can retain cellular functions and thus may regain their ability to reproduce and thereby pose a health risk. Other organisms may only exist in a symbiotic relationship with another organism such as an amoeba in their natural habitat and thus may be undetectable in a standard culture-based test.
Methods based on culture will not detect viable, non-culturable bacteria, yet even if a bacterium can be cultured, it may only grow in specific conditions, within an optimum range of temperature, pH, osmotic conditions, and in the presence of the correct nutrients. Sub-optimal culture conditions may lead to false negatives in testing, with bacteria present in a sample unable to grow.
It is also important to match the conditions for the culture with the purpose of the test. If testing for bacteria that might directly affect human health – for example, the presence of contamination in a drug product – then it is appropriate to incubate at body temperature. But to check for the presence of contamination in stored products, the ability of those bacteria to grow at room temperature, or even under refrigeration conditions, should be considered if spoilage is to be avoided.
The pharmaceutical industry routinely uses Tryptic Soy Agar (TSA) as a culture medium with incubation at 37°C, but this tends to detect fewer bacteria than using either the Reasoner's 2A agar (R2A) medium and incubation at 22°C, or yeast extract agar at either temperature. Waterborne bacteria generally prefer lower temperatures and lower levels of nutrients. Choosing the right conditions for the culture are essential.
Analytical techniques
While there are significant issues with culture methods, none of the newer analytical techniques is perfect either. The results are always dependent on the method of detection and the viability marker that is being used. Different markers measure different things, and the selection of an alternative method to culture should take the sample type and the number of anticipated bacteria present into consideration.
Fluorescence microscopy represents the most common method used to check for the presence of viable but non-culturable (VBNC) bacteria, but in some studies, culture-based methods gave higher counts than microscopic techniques. The implication is that some of the bacteria are being lost during sample preparation, or that the staining method is missing them. The different visualisation techniques, including nucleic acid stains, acridine orange, and respiratory marker, 5-cyano-2,3-ditolyl tetrazolium chloride (CTC), can give completely different results on matched pairs of samples.
Furthermore, these markers can be poor at discriminating between viable and dead bacteria after disinfection, particularly where UV is the disinfection mechanism (see Figure 1).
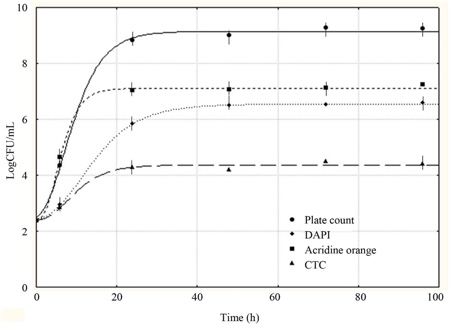
Figure 1: The relative sensitivity of some microscopic methods
Solid phase scanning uses a laser to count bacteria. Litre-sized samples are not uncommon, as the sample is filtered, and it is the bacteria present on the surface of the filter that are counted. While this technique generally detects more bacteria than normal culture techniques, there is no consistent relationship between the number of organisms detected by culture or by laser scanning. Differences between the two procedures are largely due to the sample source together with the type of culture used and the mechanism of visualising the bacteria during laser scanning.
Flow cytometry is another technique that can be applied to the enumeration of bacteria in water. The technique typically uses a sample size of the order of 100µl. It is therefore suitable only for the examination of samples with relatively high microbial counts such as potable water. As with other techniques for the detection of VBNC, flow cytometry will identify the presence of all bacteria within the sample (depending on the fluorescent marker being used), whether they are culturable or not.
Polymerase chain reaction (PCR) is capable of detecting anything that contains DNA, viable or not, and intercalating dyes can be combined with the technique as the dye assists in pinpointing nonviable bacteria. If the membrane of a bacterial cell has been damaged, perhaps by a disinfection process, the dye can enter the cell and intercalate between the DNA bases. This stops the PCR process in its tracks, and thus only viable bacteria, with no dye inserted into the nucleic acid, are amplified.
The effectiveness of the dye intercalation procedure appears to be dependent on the size of the amplicon (the genetic fragment that is being amplified). The larger the amplicon size, the more effective dye intercalation is in distinguishing between viable and non-viable bacteria.
Combination techniques
Legionella is an important organism to test for, particularly in hot water systems, but there are significant problems with its detection. While there are more than 40 species of Legionella, Legionella pneumophila is considered to be the organism responsible for the majority of human infections. Their natural habitat is water, with a particular tendency to colonise man-made structures. Some are capable of co-existing with or parasitising amoebae which affords them protection from residual disinfectant.
Detection is usually carried out via culture or PCR techniques, but neither is ideal. For culture, the incubation period is particularly lengthy, at 7–10 days, and the sensitivity is poor because of the presence of competing bacteria and the need for very selective procedures.
The culture procedure is designed to detect Legionella species, of which there are 40. However, in practice only a handful of species are efficiently recovered and further confirmation steps are needed at the end of the incubation period. PCR techniques are not ideal, either, as they also detect dead cells, and there are questions about primer specificity, particularly with primers directed at detecting the whole genus.
One suggested way around these issues is to combine culture and PCR techniques. To check the success of this strategy, three samples were first concentrated via filtration. One filter was placed into liquid enrichment and cultured onto GPVC polymyxin*, the second co-cultured with amoeba, and the third subjected to a standard GPVC culture. By far the highest results were seen via PCR on the co-culture after seven days; this was the only way that non-L. pneumophila legionella species were detected in any appreciable numbers (see Table 1).
| Table 1: Recovery of Legionella from 40 samples | |||
| Method | 2007 | L. pneumophila | Legionella species |
| Culture | 12 | 1 | |
| Enrichment PCR | 2 days | 2 | 0 |
| Enrichment PCR | 5 days | 5 | 0 |
| Enrichment PCR | 7 days | 23 | 0 |
| Co-culture PCR | 2 days | 3 | 0 |
| Co-culture PCR | 5 days | 9 | 5 |
| Co-culture PCR | 7 days | 32 | 26 |
This makes two important points. First, overall culture procedures are fairly poor, and relying on these alone is not an effective way of detecting Legionella species. Second, it is interesting to note that although Legionella are waterborne bacteria, the only species that grew well in liquid media in this study was L. pneumophila. Other Legionella species are much less likely to grow in liquid enrichment conditions, despite their presence being clear in those samples subjected to co-culture conditions. The explanation is probably that in water systems they survive either in amoebae or in biofilms on the sides of the pipes.
Medium type and nature
For some bacteria such as Pseudomonas aeruginosa, whether culture is carried out via liquid or solid media can make a big difference, as can the make-up of the medium. To investigate this further, three parallel tests were run on matching samples, prepared either via membrane filtration, liquid enrichment or using the low-nutrient liquid-based Pseudalert test. Membrane filtration found fewest bacteria. More were found using liquid enrichment, but by far the largest number were detected using the low nutrient system (see Table 2).
| Table 2: Recovery of Pseudomonas aeruginosa (142 samples) | ||
| Method | Presumptive | Confirmed |
| Membrane filtration | 18 | 16 |
| MPN | 28 | 23 |
| Pseudalert | 45 | 42 |
E.coli and coliforms represent another of the bacterial groups that it is particularly important to test for, but these are particularly sensitive to culture conditions. With the three contrasting types of tests described above, membrane filtration onto a solid medium again detected fewest bacteria, while liquid enrichment with the same medium detected more. In contrast, the low nutrient medium, Colilert, detected the highest number of both E.coli and total coliforms. Low nutrient levels would appear to be crucial in their successful detection (see Table 3).
| Table 3: Recovery of E.coli and total coliforms | ||
| Method | E.coli | Total coliforms |
| Membrane filtration (MLSA) | 16 | 43 |
| MPN (MLSB) | 22 | 49 |
| Colilert | 27 | 72 |
The medium of choice also matters when carrying out tests for heterotrophic plate counts (HPC). These tests are designed to pick up all heterotrophic organisms that rely upon organic sources of carbon to grow. Despite being routinely used to test drinking water, the plate-based HPC method counts only a relatively small proportion of the viable bacteria, and thus can miss a significant proportion of the bacteria present.
Again, using a low nutrient, liquid-based system, the HPC for QuantiTray, can detect a much greater proportion of the organisms present in a sample. This method avoids the shock of filtration, and growth is detected using fluorochrome cleavage.
In all of these cases, the low-nutrient liquid-based system gives better detection levels. It is likely that this is because it puts the bacteria under less physical stress, particularly when compared with the shock presented by the filtration process, and the exposure to the atmosphere that occurs when culturing bacteria on a solid medium. Yet despite being a process that greatly increases the likelihood of false negatives, membrane filtration continues to dominate in the water testing market.
Bacteria do not grow as well on solid media, so it is advisable to shift from agar plates to liquid cultures if possible
Bacteria do not grow as well on solid media, so it is advisable to shift from agar plates to liquid cultures if possible. As well as the shock of the filtration process, it is likely that the vastly higher concentration of oxygen to which a plate is exposed also affects growth potential, particularly for water-borne bacteria, which have evolved to thrive in the relatively low oxygen environment in water. Similarly, such bacteria normally exist in a low nutrient environment, so it is perhaps unsurprising that these are the conditions in which they grow best. It also appears to be even more important to use liquid media when testing after disinfection procedures as they are significantly better at detecting damaged bacteria.
Overall, it is essential that the testing protocol and culture conditions are optimised for the bacteria in question. Relying dogmatically on a single method for all water tests runs the risk of missing bacterial contamination and underestimating counts, regardless of whether the bacteria are culturable or nonculturable.
* GPVC polymixin is an agar containing cycloheximide glycine, vancomycin and antibiotics
Pseudalert Colilert and QuantiTray are trade names registered to IDEXX Laboratories
